Calculating effect sizes
Calculating effect sizes
Load packages
require(gdata)
require(metafor)
require(dplyr)
require(ggplot2)
Download data
For this exercise, we’ll use the Curtis et al. (1999) data set that contains an array of plant measurements taken on 65 tree species under control and elevated CO2 levels This data has been extracted from 84 papers and includes mean, standard deviation, coefficient of variation, and number of observations.
curtis<-read.xls("http://www.nceas.ucsb.edu/meta/Curtis/Curtis_CO2_database.xls",as.is=TRUE,verbose=FALSE,sheet=1)
str(curtis)
## 'data.frame': 784 obs. of 29 variables:
## $ OBSNO : int 1 2 3 4 5 6 7 8 9 10 ...
## $ PAP_NO : int 44 44 44 44 44 44 44 44 44 44 ...
## $ PARAM : chr "PN" "PN" "AGWT" "AGWT" ...
## $ P_UNIT : chr "umolCO2/m2/s" "umolCO2/m2/s" "mg" "mg" ...
## $ GENUS : chr "ALNUS" "ALNUS" "ALNUS" "ALNUS" ...
## $ SPECIES : chr "RUBRA" "RUBRA" "RUBRA" "RUBRA" ...
## $ DIV1 : chr "WOODY" "WOODY" "WOODY" "WOODY" ...
## $ DIV2 : chr "N2FIX" "N2FIX" "N2FIX" "N2FIX" ...
## $ AMBC : num 350 350 350 350 350 350 350 350 350 350 ...
## $ ELEV : num 650 650 650 650 650 650 650 650 650 650 ...
## $ CO2_UNIT: chr "ul/l" "ul/l" "ul/l" "ul/l" ...
## $ TIME : int 46 46 47 47 47 47 47 47 47 47 ...
## $ POT : chr "0.5" "0.5" "0.5" "0.5" ...
## $ METHOD : chr "GC" "GC" "GC" "GC" ...
## $ STOCK : chr "SEED" "SEED" "SEED" "SEED" ...
## $ XTRT : chr "FERT" "FERT" "FERT" "FERT" ...
## $ LEVEL : chr "HI" "CONTROL" "HI" "CONTROL" ...
## $ QUANT : chr "20mgN/l" "." "20mgN/l" "." ...
## $ SOURCE : chr "T3" "T3" "T1" "T1" ...
## $ X_AMB : num 11.8 11.7 2500.5 1460.5 1444.5 ...
## $ SE_AMB : num 0.64 1.16 324.3 76.6 219.6 ...
## $ SD_AMB : num 1.43 2.59 725.16 171.28 491.04 ...
## $ CV._AMB : num 12.8 23.3 30.5 12.3 35.7 ...
## $ N_AMB : int 5 5 5 5 5 5 5 5 5 5 ...
## $ X_ELEV : num 23.2 25.9 4076.4 1805.3 2740.6 ...
## $ SE_ELEV : num 4.61 1.48 601.7 221.1 525.2 ...
## $ SD_ELEV : num 10.31 3.31 1042.17 494.39 909.67 ...
## $ CV._ELEV: num 46.7 13.4 27.7 28.8 36 ...
## $ N_ELEV : int 5 5 3 5 3 5 3 5 3 5 ...
Please note that you may have to specify the location of the ‘perl interpreter’, e.g.
perl<-"http://www.nceas.ucsb.edu/meta/Curtis"
curtis<-read.xls("http://www.nceas.ucsb.edu/meta/Curtis/Curtis_CO2_database.xls",as.is=TRUE,verbose=FALSE,sheet=1,perl=perl)
Calculate effect sizes
For the Curtis data set, the response variables are all measured on a continuous scale. This allows us to calculate a number of traditional effect sizes, including Hedge’s D and the log-response ratio. In ‘metafor’ these effect sizes are referred to as ‘SMD’ and ‘ROM’, respectively.
In this case, we need the following variables to calculate effect sizes: mean, standard deviation, and sample size.
Additionally, sampling variances can be estimated differently. The default setting (used here) is the ‘large-sample approximation’ (vtype=’LS’). For Hedge’s D, one can also estimate ‘unbiased estimates’ (vtype=’UB’); for log-response ratio, ‘LS’ calculates sampling variance without assuming homoscedasticity while ‘HO’ assumes that variances are the same in control and treatment groups.
Note that ‘group 1’ (e.g. ‘m1i’ , ‘sd1i’) correspond to the treatment group and ‘group 2’ (e.g. ‘m2i’, ‘sd2i’) correspond to the control group.
Hedges’ D: Photosynthesis
curtis_ES<-escalc(measure='SMD', m2i=X_AMB , sd2i=SD_AMB, n2i=N_AMB, m1i=X_ELEV, sd1i=SD_ELEV, n1i=N_ELEV, vtype='LS',var.names=c("Hedges_D","Hedges_var"),data=curtis)
Visualize effect sizes (part I): forest plots
Please note that the first 25 observations are presented for visualization reasons. If you want to look at all effect sizes, use the name of the larger data frame (‘hedges_PN’) in the ‘forest’ function.
hedges_PN<-filter(curtis_ES, PARAM=="PN")
hedges_PN<-arrange(hedges_PN, GENUS)
hedges_PNN<-hedges_PN[1:25,] # not necessary
forest(hedges_PNN$Hedges_D,hedges_PNN$Hedges_var, slab=hedges_PNN$GENUS, showweights=FALSE)
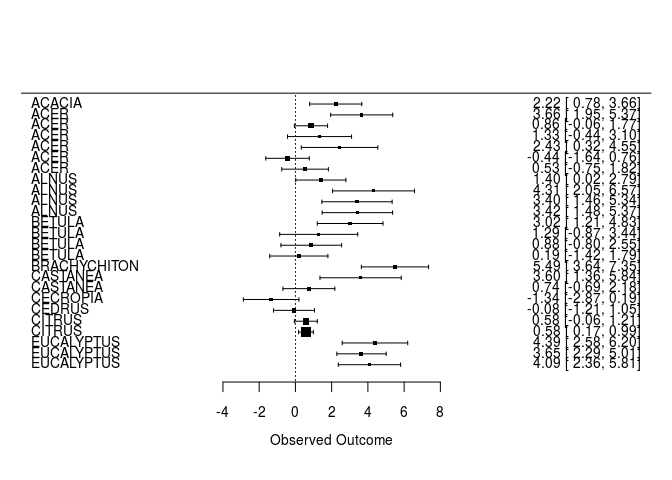
QUESTION: Do trees have higher or lower photosynthesis under ambient or elevated CO2 conditions?
Visualize effect sizes (part II): explore moderators
Alternatives for visually assessing effect sizes (per group) include histograms or density distributions.
hedges_PN$SE<-sqrt(hedges_PN$Hedges_var)
dodge <- position_dodge(width=1)
ggplot(hedges_PN, aes(x=DIV2, y=Hedges_D, colour=DIV2)) +
geom_hline(yintercept=0,color="red")+
#geom_errorbar(aes(ymin=Hedges_D-SE, ymax=Hedges_D+SE), width=.1,position=dodge) +
geom_point(position=dodge)+ labs(x="Plant group", y="Effect size")+
guides(fill=FALSE,colour=guide_legend(title="Plant group",title.position = "top"))+
theme_bw()
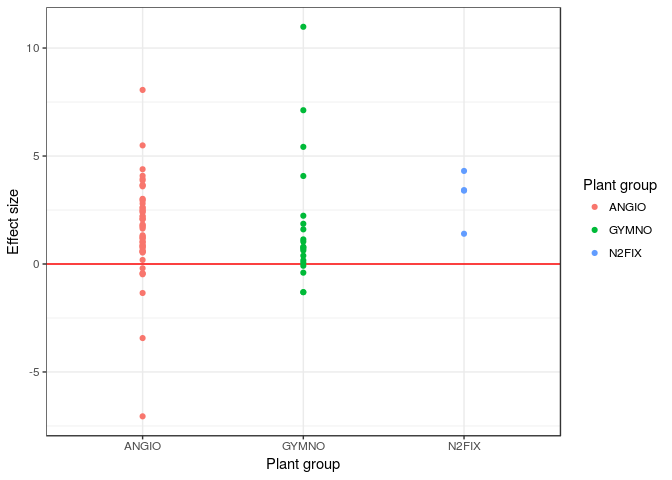
QUESTION: Are there differences among functional groups in terms of their response to elevated CO2?#
LRR: water-use efficiency
The log-response ratio is a commonly used effect size and – once back-transformed – is easier to understand (e.g. the mean effect size is 10% greater than the control).
require(metafor,quietly = TRUE)
curtis_ES2<-escalc(measure='ROM', m2i=X_AMB , sd2i=SD_AMB, n2i=N_AMB, m1i=X_ELEV, sd1i=SD_ELEV, n1i=N_ELEV, vtype='LS',var.names=c("LRR","LRR_var"),data=curtis)
## Warning in log(m1i/m2i): NaNs produced
str(curtis_ES2)
## Classes 'escalc' and 'data.frame': 784 obs. of 31 variables:
## $ OBSNO : int 1 2 3 4 5 6 7 8 9 10 ...
## $ PAP_NO : int 44 44 44 44 44 44 44 44 44 44 ...
## $ PARAM : chr "PN" "PN" "AGWT" "AGWT" ...
## $ P_UNIT : chr "umolCO2/m2/s" "umolCO2/m2/s" "mg" "mg" ...
## $ GENUS : chr "ALNUS" "ALNUS" "ALNUS" "ALNUS" ...
## $ SPECIES : chr "RUBRA" "RUBRA" "RUBRA" "RUBRA" ...
## $ DIV1 : chr "WOODY" "WOODY" "WOODY" "WOODY" ...
## $ DIV2 : chr "N2FIX" "N2FIX" "N2FIX" "N2FIX" ...
## $ AMBC : num 350 350 350 350 350 350 350 350 350 350 ...
## $ ELEV : num 650 650 650 650 650 650 650 650 650 650 ...
## $ CO2_UNIT: chr "ul/l" "ul/l" "ul/l" "ul/l" ...
## $ TIME : int 46 46 47 47 47 47 47 47 47 47 ...
## $ POT : chr "0.5" "0.5" "0.5" "0.5" ...
## $ METHOD : chr "GC" "GC" "GC" "GC" ...
## $ STOCK : chr "SEED" "SEED" "SEED" "SEED" ...
## $ XTRT : chr "FERT" "FERT" "FERT" "FERT" ...
## $ LEVEL : chr "HI" "CONTROL" "HI" "CONTROL" ...
## $ QUANT : chr "20mgN/l" "." "20mgN/l" "." ...
## $ SOURCE : chr "T3" "T3" "T1" "T1" ...
## $ X_AMB : num 11.8 11.7 2500.5 1460.5 1444.5 ...
## $ SE_AMB : num 0.64 1.16 324.3 76.6 219.6 ...
## $ SD_AMB : num 1.43 2.59 725.16 171.28 491.04 ...
## $ CV._AMB : num 12.8 23.3 30.5 12.3 35.7 ...
## $ N_AMB : int 5 5 5 5 5 5 5 5 5 5 ...
## $ X_ELEV : num 23.2 25.9 4076.4 1805.3 2740.6 ...
## $ SE_ELEV : num 4.61 1.48 601.7 221.1 525.2 ...
## $ SD_ELEV : num 10.31 3.31 1042.17 494.39 909.67 ...
## $ CV._ELEV: num 46.7 13.4 27.7 28.8 36 ...
## $ N_ELEV : int 5 5 3 5 3 5 3 5 3 5 ...
## $ LRR : atomic 0.679 0.795 0.489 0.212 0.64 ...
## ..- attr(*, "measure")= chr "ROM"
## ..- attr(*, "ni")= int 10 10 8 10 8 10 8 10 8 10 ...
## $ LRR_var : num 0.0424 0.0131 0.0386 0.0178 0.0598 ...
## - attr(*, "digits")= num 4
## - attr(*, "yi.names")= chr "LRR"
## - attr(*, "vi.names")= chr "LRR_var"
Visualize effect sizes (part I) : forest plots
require(metafor,quietly = TRUE)
require(dplyr,quietly=TRUE)
LRR_WUE<-filter(curtis_ES2, PARAM=="WUE")
LRR_WUE<-arrange(LRR_WUE, GENUS)
forest(LRR_WUE$LRR,LRR_WUE$LRR_var, slab=LRR_WUE$GENUS, showweights=TRUE)
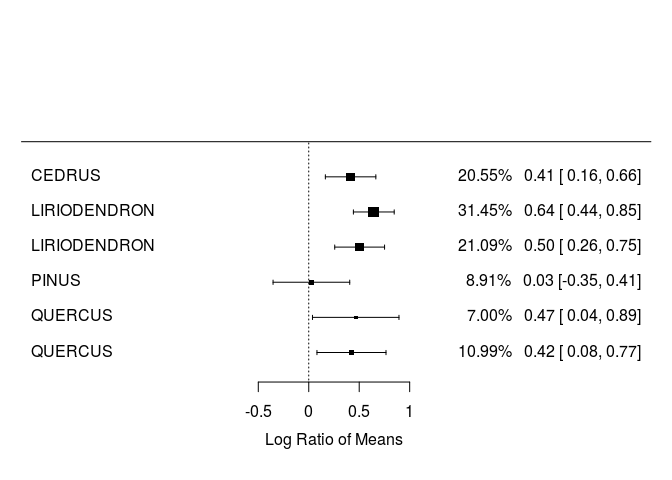
Note that the percentage next to each effect size is the weight of each point.
QUESTION: Do trees have higher or lower water-use efficiency in response to elevated CO2?