Hierarchical meta-analytical models
Hierarchical meta-analytical models
Getting started
Load packages
require(gdata)
require(metafor)
require(dplyr)
require(multcomp)
require(ggplot2)
Download data (Curtis et al. 1999)
curtis<-read.xls("http://www.nceas.ucsb.edu/meta/Curtis/Curtis_CO2_database.xls",as.is=TRUE,verbose=FALSE,sheet=1)
curtis_ES<-escalc(measure='ROM', m2i=X_AMB , sd2i=SD_AMB, n2i=N_AMB, m1i=X_ELEV, sd1i=SD_ELEV, n1i=N_ELEV, vtype='LS',var.names=c("LRR","LRR_var"),data=curtis)
## Warning in log(m1i/m2i): NaNs produced
summary(as.factor(curtis_ES$PARAM))
## AGWT BD BGWT CRWT FRWT GS GS_AC HT INDLA JMAX
## 14 25 64 11 9 49 12 30 3 3
## LAR LFC LFNA LFNM LFP LFSTAR LFSUG LFTNC LFWT MAXLA
## 15 3 15 42 5 19 11 7 50 41
## PIRC PN PN_AC RD RD_AC RGR SEEDWT SLA SLW STWT
## 2 79 29 17 3 17 1 18 15 47
## TOTN TOTWT VCMAX WUE WUE_AC
## 9 102 9 6 2
curtis_WT<-filter(curtis_ES, PARAM=="TOTWT") # let's use whole plant weight because it has the largest number of observations
curtis_WT$GEN_SPP<-paste(curtis_WT$GENUS,curtis_WT$SPECIES,sep="_")
Part I: compare fixed and random effects models
Fixed effects model
Important Fixed effects models assume that there is one true effect size, i.e. differences are largely due to sampling error.
fix_wt<-rma(LRR, LRR_var, method="FE", data=curtis_WT)
summary(fix_wt)
##
## Fixed-Effects Model (k = 102)
##
## logLik deviance AIC BIC AICc
## -245.9580 769.0185 493.9160 496.5410 493.9560
##
## Test for Heterogeneity:
## Q(df = 101) = 769.0185, p-val < .0001
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## 0.2088 0.0054 38.3374 <.0001 0.1982 0.2195 ***
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
This model estimates the ‘grand mean’ of the effect of CO2 exposure on total plant weight.
Random effects model
Important Random effects models allows the true effect size to differ. Here, the effect sizes represent a random sample from a particular distribution.
You can use either the ‘rma’ function, which uses REML as the default method for fitting a model, or ‘rma.mv’, which requires that you specify the random effects.
Here, we use a random group term for each study (‘PAP_NO’) because many studies report more than one effect size.
## [1] 3.517241
re_wt<-rma.mv(LRR, LRR_var, random=~1|PAP_NO, data=curtis_WT)
summary(re_wt)
##
## Multivariate Meta-Analysis Model (k = 102; method: REML)
##
## logLik Deviance AIC BIC AICc
## -50.0285 100.0570 104.0570 109.2872 104.1794
##
## Variance Components:
##
## estim sqrt nlvls fixed factor
## sigma^2 0.0147 0.1212 29 no PAP_NO
##
## Test for Heterogeneity:
## Q(df = 101) = 769.0185, p-val < .0001
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## 0.2603 0.0267 9.7528 <.0001 0.2080 0.3127 ***
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
This model also estimates the ‘grand mean’ of the effect of CO2 exposure on total plant weight.
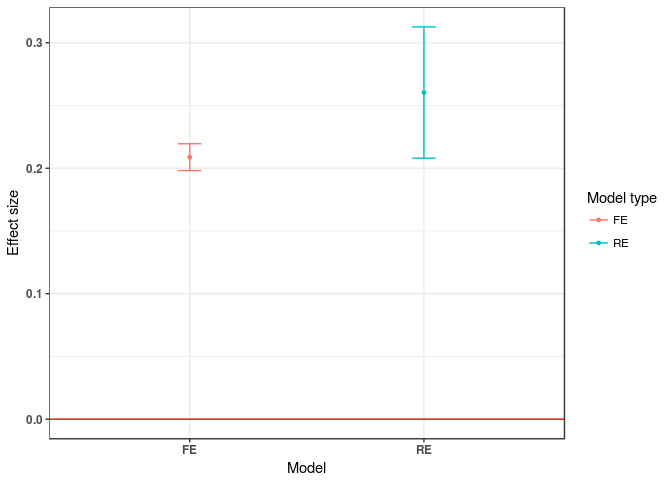
Comparison of model heterogeneity
Note that I2 for the random effects model integrates heterogeneity from within- and between-clusters following Nakagawa & Santos (2012).
See here to calculate I2 for between- and within-clusters of multilevel models separately.
I2_fe<-cbind.data.frame("model"="fixed","I2"=fix_wt$I2)
#######################################
# calcualte I2 for a multilevel model #
#######################################
W <- diag(1/curtis_WT$LRR_var)
X <- model.matrix(re_wt)
P <- W - W %*% X %*% solve(t(X) %*% W %*% X) %*% t(X) %*% W
re_I2<-100 * sum(re_wt$sigma2) / (sum(re_wt$sigma2) + (re_wt$k-re_wt$p)/sum(diag(P)))
I2_re<-cbind.data.frame("model"="random","I2"=re_I2)
#######################################
I22<-rbind.data.frame(I2_fe,I2_re)
I22
## model I2
## 1 fixed 86.86638
## 2 random 81.76727
Part II: Hierarchical, multi-level model (‘meta-regression’)
Identify most parsimonious random-effects structure
I’ve included additional random effects included in the data set: extra treatment (e.g. nutrient addition, light, water), species identity, and a combination of the two.
re_wt1<-rma.mv(LRR, LRR_var, mods=~DIV2,random=~1|PAP_NO, data=curtis_WT)
re_wt2<-rma.mv(LRR, LRR_var, mods=~DIV2,random=list(~1|PAP_NO, ~1|XTRT), data=curtis_WT)
re_wt3<-rma.mv(LRR, LRR_var, mods=~DIV2,random=list(~1|PAP_NO, ~1|GEN_SPP), data=curtis_WT)
re_wt4<-rma.mv(LRR, LRR_var, mods=~DIV2,random=list(~1|PAP_NO, ~1|XTRT, ~1|GEN_SPP), data=curtis_WT)
AICc<-rbind(mod1=re_wt1$fit.stats$REML[5],mod2=re_wt2$fit.stats$REML[5],mod3=re_wt3$fit.stats$REML[5],mod4=re_wt4$fit.stats$REML[5])
AICc # and the winner is ...
## [,1]
## mod1 110.91380
## mod2 74.29536
## mod3 87.97844
## mod4 54.58859
Test significance of moderators
There are a couple of ways to test the signifance of moderator variables.
Likelihood Ratio Tests
Note that maximum likelihood is used to fit both models
bigg<-rma.mv(LRR, LRR_var, mods=~DIV2,random=list(~1|PAP_NO, ~1|XTRT, ~1|GEN_SPP), data=curtis_WT, method="ML")
small<-rma.mv(LRR, LRR_var, mods=~1,random=list(~1|PAP_NO, ~1|XTRT, ~1|GEN_SPP), data=curtis_WT, method="ML")
anova(bigg,small)
## df AIC BIC AICc logLik LRT pval QE
## Full 6 51.7179 67.4677 52.6021 -19.8589 719.5615
## Reduced 4 47.8123 58.3122 48.2247 -19.9062 0.0945 0.9539 769.0185
Knapp & Hartung Adjustment
The Knapp & Hartung adjustment, similar to the likelihood ratio test, can be used for individual model coefficients (t-tests) or a group of model coefficients (F tests).
bigg<-rma.mv(LRR, LRR_var, mods=~DIV2,random=list(~1|PAP_NO, ~1|XTRT, ~1|GEN_SPP), data=curtis_WT, test="knha")
summary(bigg)
##
## Multivariate Meta-Analysis Model (k = 102; method: REML)
##
## logLik Deviance AIC BIC AICc
## -20.8378 41.6755 53.6755 69.2463 54.5886
##
## Variance Components:
##
## estim sqrt nlvls fixed factor
## sigma^2.1 0.0104 0.1019 29 no PAP_NO
## sigma^2.2 0.0093 0.0965 8 no XTRT
## sigma^2.3 0.0042 0.0649 37 no GEN_SPP
##
## Test for Residual Heterogeneity:
## QE(df = 99) = 719.5615, p-val < .0001
##
## Test of Moderators (coefficient(s) 2:3):
## QM(df = 2) = 0.0914, p-val = 0.9553
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## intrcpt 0.2778 0.0501 5.5423 <.0001 0.1796 0.3760 ***
## DIV2GYMNO -0.0044 0.0560 -0.0788 0.9372 -0.1142 0.1054
## DIV2N2FIX 0.0481 0.1687 0.2852 0.7755 -0.2826 0.3788
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
In both cases, including plant group as a moderator variable did not add information to the model.
Model Heterogeneity (I2)
W <- diag(1/curtis_WT$LRR_var)
X <- model.matrix(bigg)
P <- W - W %*% X %*% solve(t(X) %*% W %*% X) %*% t(X) %*% W
100 * sum(bigg$sigma2) / (sum(bigg$sigma2) + (bigg$k-bigg$p)/sum(diag(P)))
## [1] 86.99147
Note that the I2 for this model is very similar to that of the fixed effects model and the random effects model with a simpler random effects structure (seen earlier).
Pairwise comparisons
We will use the ‘multcomp’ package to test contrasts between levels of the factor ‘DIV2’.
require(multcomp)
bigg_int<-rma.mv(LRR, LRR_var, mods=~DIV2-1,random=list(~1|PAP_NO, ~1|XTRT, ~1|GEN_SPP), data=curtis_WT, test="knha")
summary(bigg_int)
##
## Multivariate Meta-Analysis Model (k = 102; method: REML)
##
## logLik Deviance AIC BIC AICc
## -20.8378 41.6755 53.6755 69.2463 54.5886
##
## Variance Components:
##
## estim sqrt nlvls fixed factor
## sigma^2.1 0.0104 0.1019 29 no PAP_NO
## sigma^2.2 0.0093 0.0965 8 no XTRT
## sigma^2.3 0.0042 0.0649 37 no GEN_SPP
##
## Test for Residual Heterogeneity:
## QE(df = 99) = 719.5615, p-val < .0001
##
## Test of Moderators (coefficient(s) 1:3):
## QM(df = 3) = 36.2522, p-val < .0001
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## DIV2ANGIO 0.2778 0.0501 5.5423 <.0001 0.1796 0.3760 ***
## DIV2GYMNO 0.2734 0.0598 4.5721 <.0001 0.1562 0.3906 ***
## DIV2N2FIX 0.3259 0.1691 1.9273 0.0539 -0.0055 0.6574 .
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
summary(glht(bigg_int, linfct=rbind(c(-1,1,0), c(-1,0,1), c(0,-1,1))), test=adjusted("none"))
##
## Simultaneous Tests for General Linear Hypotheses
##
## Fit: rma.mv(yi = LRR, V = LRR_var, mods = ~DIV2 - 1, random = list(~1 |
## PAP_NO, ~1 | XTRT, ~1 | GEN_SPP), data = curtis_WT, test = "knha")
##
## Linear Hypotheses:
## Estimate Std. Error z value Pr(>|z|)
## 1 == 0 -0.004411 0.056006 -0.079 0.937
## 2 == 0 0.048123 0.168718 0.285 0.775
## 3 == 0 0.052534 0.173766 0.302 0.762
## (Adjusted p values reported -- none method)

Exercise
- download data from our Github repository
install.packages('RCurl')
require(RCurl)
landuse <- getURL("https://raw.githubusercontent.com/dylancraven/MetaAnalysis_Course/gh-pages/Slides/LandUseBiodiv.csv")
landuse <- read.csv(text = landuse,header=T)
-
Fit a multi-level model
-
Calculate I2
-
Test for multiple comparisons (if using a categorical moderator)